pandas#
Now that we’ve discussed Scientific Computing generally and introduced NumPy, we’re ready to learn the basics of pandas, the most popular python package for working with data. Similar to what we saw with NumPy, we’ll first introduce the core object at the heart of pandas – the DataFrame. From there, we’ll introduce some of the attributes and most common methods for working with DataFrames. And, we’ll finish discussing additional functions within pandas that make working with data more straightforward.
As with NumPy, you’ll need to first import pandas. Convention is to import: import padas as pd, so we’ll encourage that here. Using this import statement, any time you want to reference pandas functionality, you can do so with just the two letters pd.
pandas using import pandas as pd before attempting to run any of the included code in this section.
import pandas as pd
Heterogeneous Data#
While NumPy arrays make working with homogenous data (data that are all of one type, typically numbers), pandas adds functionality for working with heterogeneous data. What if you have information about individual’s heights (floats) but also the color of their eyes (strings)? This would be an example of heterogeneous data and is precisely what pandas is designed to handle.
More specifically, a DataFrame can store multiple types of data; however, each individual column will store a single type of data.
pandas DataFrame#
Enter, the DataFrame - the central object within pandas. DataFrames are a 2-dimensional data structure that store data in a table/array, but that enable continous (numeric) and categorical (discrete and text-based) data to be stored in a single object. DataFrames enable information to be stored in rows and columns (like an array); however, columns can be of mixed type and both rows and columns can have labels. If you’re envisioning data stored in a spreadsheet-like object, you’ve got the right idea. For example, a DataFrame could be used to store the following information:
| Participant ID | Height (cm) | Eye Color |
|---|---|---|
| 01 | 170 | Blue |
| 02 | 165 | Green |
| 03 | 180 | Brown |
| 04 | 175 | Hazel |
| 05 | 160 | Brown |
In fact, we can store that very information in a pandas DataFrame using pd.DataFrame and passing in the data:
df = pd.DataFrame({'Participant ID': ['01', '02', '03', '04', '05'],
'Height (cm)': [170, 165, 180, 175, 160],
'Eye Color': ['Blue', 'Green', 'Brown', 'Hazel', 'Brown']
})
df
| Participant ID | Height (cm) | Eye Color | |
|---|---|---|---|
| 0 | 01 | 170 | Blue |
| 1 | 02 | 165 | Green |
| 2 | 03 | 180 | Brown |
| 3 | 04 | 175 | Hazel |
| 4 | 05 | 160 | Brown |
We’ll note a few things about this df object:
We’ve called the object
df. This is a convention you’ll often see when people are working withDataFrames withdfbeing short fordf. We’ll use this convention frequently, but you could give yourDataFrames more informative names (i.e.df_participants).In
dfyou’ll notice that there are three column names at top.You’ll also notice there are indices off to the left for each row. By default,
pandaswill assign each row an index starting with zero.
Attributes#
As with arrays, when working with DataFrame objects, it’s often helpful to be able to access information about the DataFrame by accessing attributes.
shape#
To get the dimensions of the DataFrame, the attribute is the same as in Numpy: shape
df.shape
(5, 3)
Again, similar to arrays, the output specifies the number of rows first (5) and the number of columns second (3).
columns & index#
If you ever need to access the names of the columns in a DataFrame it can be helpful to know that the column names can be extracted from the columns attribute:
df.columns
Index(['Participant ID', 'Height (cm)', 'Eye Color'], dtype='object')
The same goes for extracting the indices (row names). This can be accessed through index:
df.index
RangeIndex(start=0, stop=5, step=1)
dtypes#
dtypes can also be helpful to determine how the DataFrame is storing your data. This is particularly important in pandas due to the fact that DataFrames can store heterogenous data…and pandas makes its best guess as to the information stored in the DataFrame. This process is not perfect. If you’re working with a very large dataset and you expect all of the values in a given column to be numbers, you may assume that the column stores numbers. However, what if one of the values in your large dataset was actually the word “nine feet”? In that case, pandas would convert everything to strings in that column as it doesn’t know how to handle “nine feet” as a number (even if we humans may!). This is why it’s good practice to just double check that each of your columns is of the expected type using the dtypes attribute:
df.dtypes
Participant ID object
Height (cm) int64
Eye Color object
dtype: object
In the above, you may have assumed that Participant ID would be an integer; however, despite appearing as numbers these IDs were entered as strings, making them object type variables.
head#
Also note that head returns the first five rows of a DataFrame. This can be helpful when working with larger DataFrames just to get a sene of the data.
Note that there is both a head attribute:
df.head
<bound method NDFrame.head of Participant ID Height (cm) Eye Color
0 01 170 Blue
1 02 165 Green
2 03 180 Brown
3 04 175 Hazel
4 05 160 Brown>
…and a head() method:
df.head()
| Participant ID | Height (cm) | Eye Color | |
|---|---|---|---|
| 0 | 01 | 170 | Blue |
| 1 | 02 | 165 | Green |
| 2 | 03 | 180 | Brown |
| 3 | 04 | 175 | Hazel |
| 4 | 05 | 160 | Brown |
The difference is in how the data are displayed, with the method displaying the data in a slightly more reader-friendly fasion.
Indexing & Slicing#
So far we’ve introduced the DataFrame as a way to store heterogenous data in rows and columns. But, I haven’t mentioned exactly how pandas accomplishes that. While the DataFrame is the core object, it’s also important to be aware that DataFrames are comprised of Series, which are a 1-dimensional labeled array storing data. Each column in pandas is a Series object and can be accessed using the column name and indexing. For example, if we wanted to extract the 'Height' column from our df we could do so using:
df['Height (cm)']
0 170
1 165
2 180
3 175
4 160
Name: Height (cm), dtype: int64
In the above, we can see that indexing into a DataFrame by specifying the column name, pandas will return all values in that column, along with their corresponding indices. It also reports the type of the variable and the name of the column.
Specifically, the way pandas stores each column is as a pd.Series type object. We can see this explicitly by checking the type of the above output:
type(df['Height (cm)'])
pandas.core.series.Series
Thus, conceptually, every DataFrame is comprised of Series of the same length stacked next to one another.
We’re also able to access information stored in rows; however, the syntax has to differ a little bit. df[0] will not work here, as pandas would be looking for a column with the label 0. Rather, when we want to access data in a row, we have to return to our familiar slicing syntax, first specifing the row(s) we want to return using start:stop notation.
For example, df[:1] will return from the beginning of the DataFrame up to but not including the row with the index 1 (using our familiar slicing approach):
df[:1]
| Participant ID | Height (cm) | Eye Color | |
|---|---|---|---|
| 0 | 01 | 170 | Blue |
However, we will highlight here that what is being returned here is a DataFrame (a slice of the original) and not a Series:
type(df[:1])
pandas.core.frame.DataFrame
pd.Series objects are specifically for data for a single variable in a column…and not for returning subsets of a larger DataFrame across columns. If multiple columns are involved, then it’s a DataFrame object.
If you wanted to extract the information as a series, you would need to use loc…
.loc#
But, what if you want to combine these two ideas? What if you want to specify a few rows and a few columns? This is where loc comes in. loc (standing for loccation) allows users to control which part of the DataFrame they want to return.
For example, if we wanted the second and third rows and only the Eye Color column from df we could use the following:
df.loc[1:2, 'Eye Color']
1 Green
2 Brown
Name: Eye Color, dtype: object
In the above syntax, notice that the convention of [row, column] is still followed. However, the start:stop for loc is inclusive of the stop value (unlike everywhere else in Python). Thus, to return the second and third rows, you specify that you want to start at index 1 and stop at (and include) index 2.
Returning to our goal of extracting the data in the first row from above, if we wanted to extract just the values in a given rows as a Series, we could use loc to do so, first specifiying the index and then to extract all columns:
df.loc[0,:]
Participant ID 01
Height (cm) 170
Eye Color Blue
Name: 0, dtype: object
Finally, putting all of this together, loc can also be used to extract singular values, by specifying a single location within the DataFrame:
df.loc[0, 'Eye Color']
'Blue'
Note that the above returns a value of the type of data directly, and not a DataFrame or Series. Thus, ‘Blue’ here is a string.
Methods#
There are many, many methods available for DataFrame objects. This section will only cover a few of the most commonly used, but will give you a sense of the types of things one can do with a DataFrame.
describe()#
One powerful method is the describe() method, which will calculate and provide a number of descriptive statistics for all numeric variables in your dataset. In df, we have a single numeric varaible (Height (cm)), which is summarized below:
df.describe()
| Height (cm) | |
|---|---|
| count | 5.000000 |
| mean | 170.000000 |
| std | 7.905694 |
| min | 160.000000 |
| 25% | 165.000000 |
| 50% | 170.000000 |
| 75% | 175.000000 |
| max | 180.000000 |
replace()#
Of course, DataFrames are mutable, with the ability to change or update values after creation, using replace:
For example, what if I realized that the participant IDs should be zero padded with two zeroes instead of one. To accomplish this, we can use the replace method to change each '0' into '00':
df['Participant ID'] = df['Participant ID'].replace('0', '00', regex=True)
df
| Participant ID | Height (cm) | Eye Color | |
|---|---|---|---|
| 0 | 001 | 170 | Blue |
| 1 | 002 | 165 | Green |
| 2 | 003 | 180 | Brown |
| 3 | 004 | 175 | Hazel |
| 4 | 005 | 160 | Brown |
Notice in the syntax above a few things:
The
replacemethod is operating on theSeries'Partcipant ID'. Thus, when we assign the new values, they are assigned back to that specific series. The rest ofdfremains unchanged.In
replacethe first argument is what is to be replaced, the second is what to replace it with.regex=Truespecifies to use regular expression to evaluate the change (rather than string literals). Withoutregex=True,replacewould only change a cell with the exact value ‘0’ to ‘00’…instead of changing every case where it finds a zero.
We’ll also note that it is possible to directly assign a new value to a given location within a DataFrame directly using indexing/slicing and assignment:
df.loc[0, 'Height (cm)'] = 173
df
| Participant ID | Height (cm) | Eye Color | |
|---|---|---|---|
| 0 | 001 | 173 | Blue |
| 1 | 002 | 165 | Green |
| 2 | 003 | 180 | Brown |
| 3 | 004 | 175 | Hazel |
| 4 | 005 | 160 | Brown |
astype()#
We mentioned dtypes is helpful for knowing what your value type is. But, what if you want to change it to another type. astype() allows you to typecast (meaning specify the type) for a variable. If we wanted 'Height (cm)' to be a float instead of an integer, for example, we could accomplish that using astype:
df['Height (cm)'] = df['Height (cm)'].astype(float)
df
| Participant ID | Height (cm) | Eye Color | |
|---|---|---|---|
| 0 | 001 | 173.0 | Blue |
| 1 | 002 | 165.0 | Green |
| 2 | 003 | 180.0 | Brown |
| 3 | 004 | 175.0 | Hazel |
| 4 | 005 | 160.0 | Brown |
Now instead of integers, the heights are floats.
We’ll note here that there are additional methods that help with typecasting, and we’ll mention two here:
convert_dtypes| will convert columns to the best possibledtypesto_numpy| will convert aDataFrameto aNumPyarray
value_counts()#
Often when working with data (particularly categorical data), we’re interest in how many different values there are. For example, how many different eye colors are in our dataset? And how many individuals are there with each eye color? While we can easily determine this for our limited dataset here, as datasets grow in size, it’s less easy to determine at a glance. Instead, we can use value_counts to answer this question:
df['Eye Color'].value_counts()
Eye Color
Brown 2
Blue 1
Green 1
Hazel 1
Name: count, dtype: int64
In the above output, each unique eye color is at left and the corresponding number of how many times each shows up is at right. Eye colors are sorted by frequency of appearance, with most frequent first. For categories with ties, labels are sorted alphanumerically.
unique & nunique#
Similarly, if you don’t want all of the information above, but want pieces of information about uniqueness, there are additional helpful methods.
For example unique returns all the unique values (as an array):
df['Eye Color'].unique()
array(['Blue', 'Green', 'Brown', 'Hazel'], dtype=object)
…and nunique() returns how many unique values there are (in our case, 4):
df['Eye Color'].nunique()
4
Method Chaining#
Building on the above, one key important features within pandas is the ability to chain methods together using the general syntax df.method1().method2(). In this, method1 would be applied to df and then method2() would be appled to the output of method1().
For example, what if you didn’t want the unique eye colors as an array…but rather as a list. You can do that with method chaining:
df['Eye Color'].unique().tolist()
['Blue', 'Green', 'Brown', 'Hazel']
…and what if you didn’t want all of the value_counts output…but just the category that shows up the most? You can use value_counts() chained with max(), the output of which lets you know of all the eye colors, the one that shows up the most shows up twice:
df['Eye Color'].value_counts().max()
2
…but what if you didn’t want to know how many times the most common eye color showed up, but just what that eye color that showed up the most was. There’s idxmax() for that, which can be chained with value_counts():
df['Eye Color'].value_counts().idxmax()
'Brown'
Note that method chaining is not limited to two methods. Many methods can be chained together the output of each subsequent method becoming the input for the next. The reason this is possible is because most pandas methods return a DataFrame…enabling the next method to operate on that returned DataFrame.
While we won’t walk through examples of all of the existing methods in pandas, we’ll summarize a few additional common ones here:
Function |
Purpose |
|---|---|
|
Fill in missing data |
|
Group data by a variable prior to carrying out some operation within group |
|
Combine (concatenate) |
|
Enable SQL-like joins across |
|
Create new columns |
|
Filter rows using Boolean logic |
|
Returns rows with match |
Additional methods are summarized in the pandas Cheat Sheet.
Functions#
While the DataFrame and Series objects are at the center of the pandas package, there are additional functions within pandas that support working with data in Python.
read_*#
In this chapter so far, we’ve worked with data that we generated directly and stored in a DataFrame. However, typically data live elsewhere and individuals want to read it into python for data analysis. This is where the suite of read_* functions come into play. There are a number of them, the most common of which is likely read_csv(), which reads data from CSV (comma-separated value files) into python. Files read in can be either filepaths or URLs.
For example, the following reads in data from a CSV stored on GitHub (passing in the URL of the file as a string:
df_msleep = pd.read_csv('https://raw.githubusercontent.com/ShanEllis/datasets/master/msleep.csv')
df_msleep
| name | genus | vore | order | conservation | sleep_total | sleep_rem | sleep_cycle | awake | brainwt | bodywt | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | Cheetah | Acinonyx | carni | Carnivora | lc | 12.1 | NaN | NaN | 11.9 | NaN | 50.000 |
| 1 | Owl monkey | Aotus | omni | Primates | NaN | 17.0 | 1.8 | NaN | 7.0 | 0.01550 | 0.480 |
| 2 | Mountain beaver | Aplodontia | herbi | Rodentia | nt | 14.4 | 2.4 | NaN | 9.6 | NaN | 1.350 |
| 3 | Greater short-tailed shrew | Blarina | omni | Soricomorpha | lc | 14.9 | 2.3 | 0.133333 | 9.1 | 0.00029 | 0.019 |
| 4 | Cow | Bos | herbi | Artiodactyla | domesticated | 4.0 | 0.7 | 0.666667 | 20.0 | 0.42300 | 600.000 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 78 | Tree shrew | Tupaia | omni | Scandentia | NaN | 8.9 | 2.6 | 0.233333 | 15.1 | 0.00250 | 0.104 |
| 79 | Bottle-nosed dolphin | Tursiops | carni | Cetacea | NaN | 5.2 | NaN | NaN | 18.8 | NaN | 173.330 |
| 80 | Genet | Genetta | carni | Carnivora | NaN | 6.3 | 1.3 | NaN | 17.7 | 0.01750 | 2.000 |
| 81 | Arctic fox | Vulpes | carni | Carnivora | NaN | 12.5 | NaN | NaN | 11.5 | 0.04450 | 3.380 |
| 82 | Red fox | Vulpes | carni | Carnivora | NaN | 9.8 | 2.4 | 0.350000 | 14.2 | 0.05040 | 4.230 |
83 rows × 11 columns
In addition to reading in CSV files, there are pd.read_* functions for reading in JSON, XML, HTML, and excel datasets (among others).
Similarly there are corresponding to_* methods that allow for DataFrames to be written out to files of various types (i.e. CSV, JSON, etc.)
Plotting#
While there are many ways to generate plots in python, pandas does have the ability to generate plots via the plotting module in pandas:
pd.plotting.boxplot(df_msleep, column='sleep_total', by='vore')
<Axes: title={'center': 'sleep_total'}, xlabel='vore'>
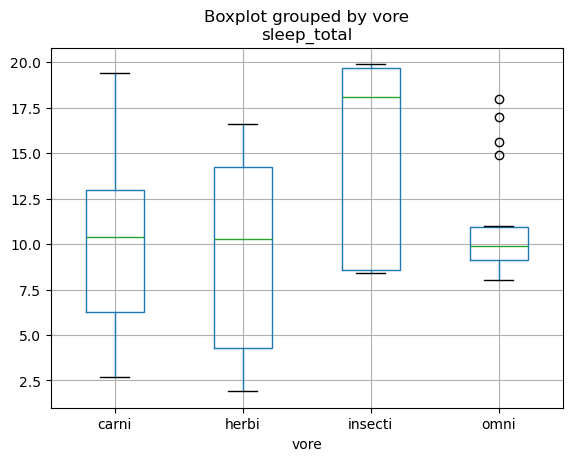
While we won’t be discussing the full capabilities here (partially because it’s beyond the scope and partially because many people use other approaches for plotting in python), we did at least want to introduce this as a general capability within pandas.
Exercises#
Q1. Comparing them to standard library Python types, which is the best mapping for NumPy arrays and pandas DataFrames?
A) DataFrames are like lists, arrays are like tuples
B) DataFrames and arrays are like lists
C) DataFrames are like tuples, arrays are like lists
D) DataFrames and arrays are like dictionaries
E) Dataframes are like dictionaries, arrays are like lists
Q2. If a DataFrame contained only numeric values, how would it differ from a NumPy array?
A) The DataFrame would additionally have indices and column names
B) They would be exactly the same
C) DataFrames cannot store homogenous data
D) The DataFrame would convert all of the numeric values to strings
E) NumPy arrays cannot store numeric values
Q3. Read in the df_msleep DataFrame provided above. Use pandas attributes to determine the dimensions and types of information stored in df_msleep
Q4. Again using df_msleep, use a pandas method to determine the average amount of total sleep the animals in this dataset get.
Q5. Again using df_msleep, use a pandas method to determine which 'vore' is most common in this dataset, as well as how many animals there are of that 'vore'.

